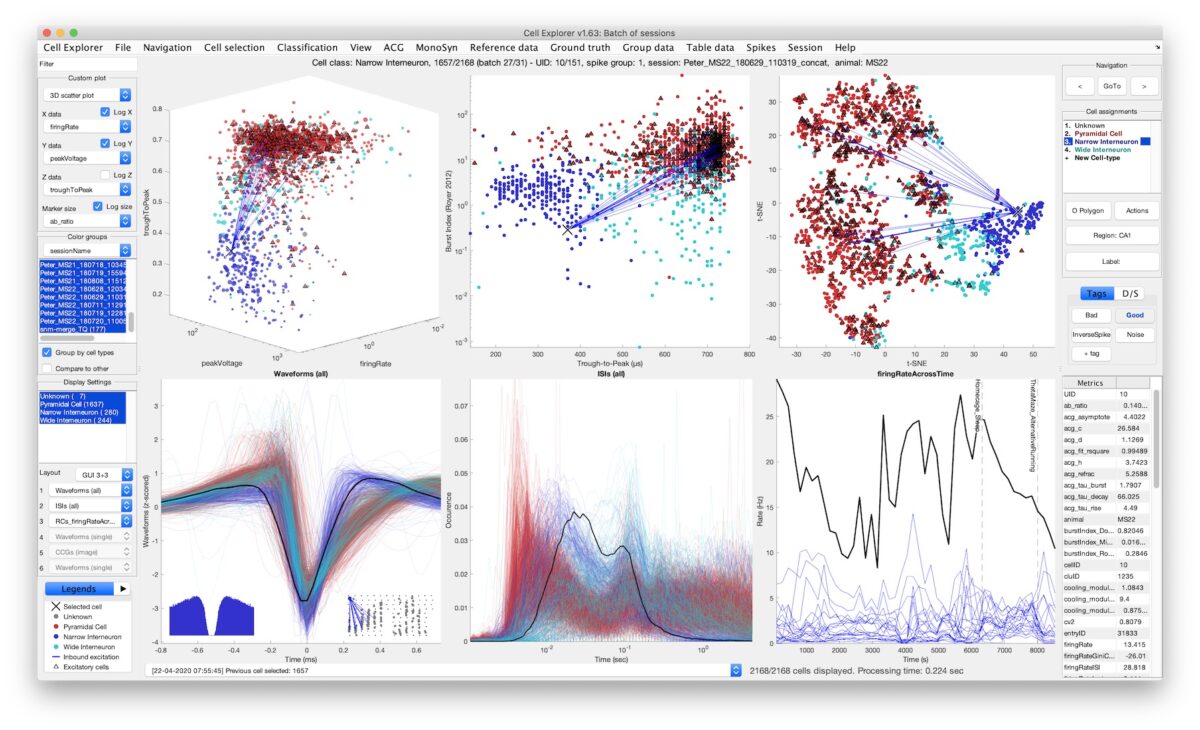
Framework for single-cell classification
The large diversity of cell types of the brain provides the means by which circuits perform complex operations. Understanding such diversity is one of the key challenges of modern neuroscience. These cells have many unique electrophysiological and behavioral features from which parallel cell types classification can be inferred. The CellExplorer is a framework for analyzing and characterizing single cells recorded using extracellular electrodes. A high dimensional representation is built from electrophysiological and functional features including the spike waveform, spiking statistics, behavioral spiking dynamics, spatial firing maps, and various brain rhythms. Moreover, we are incorporating opto-tagget cells in this pipeline (ground-truth cell types). The user-friendly graphical interface allows for verification, classification, and exploration of those same features. The framework is built entirely in Matlab making it fast and intuitive to implement your own code and incorporate the CellExplorer in your overall pipeline and analysis scripts.
Learn more about the CellExplorer at the dedicated website: CellExplorer.org
Peter C Petersen, György Buzsáki (2020). CellExplorer: a graphical user interface and standardized pipeline for visualizing and characterizing single neuron features. bioRxiv 2020.05.07.083436; doi: https://doi.org/10.1101/2020.05.07.083436. [github]